March 04, 2021
Converting nested XML to dataframe in R - a tidyverse approach
798 words | 4-min read |
TL;DR
tidyr (in tidyverse) provides functions unnest_wider and unnest_longer to transform XML data into dataframe quickly, using the same ideology of pivot_wider and pivot_longer in dplyr.
I rarely deal with XML data. However, sometimes the government discloses open data in XML format only. Currently I need to find data about the restaurant licenses in Hong Kong, and FEHD provides an open dataset. Unluckily, the dataset is available in XML format only.
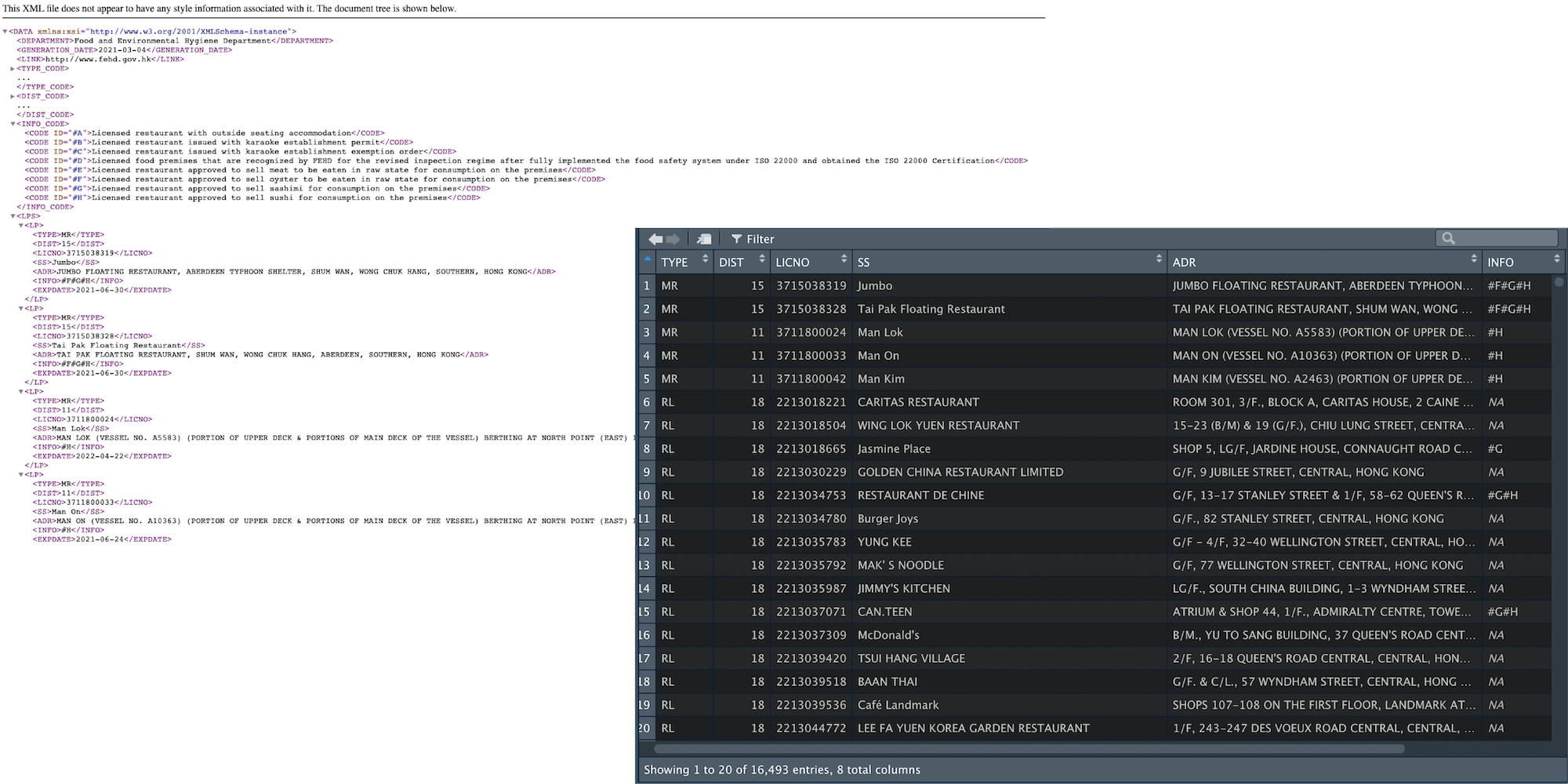
The XML data file looks like something below:
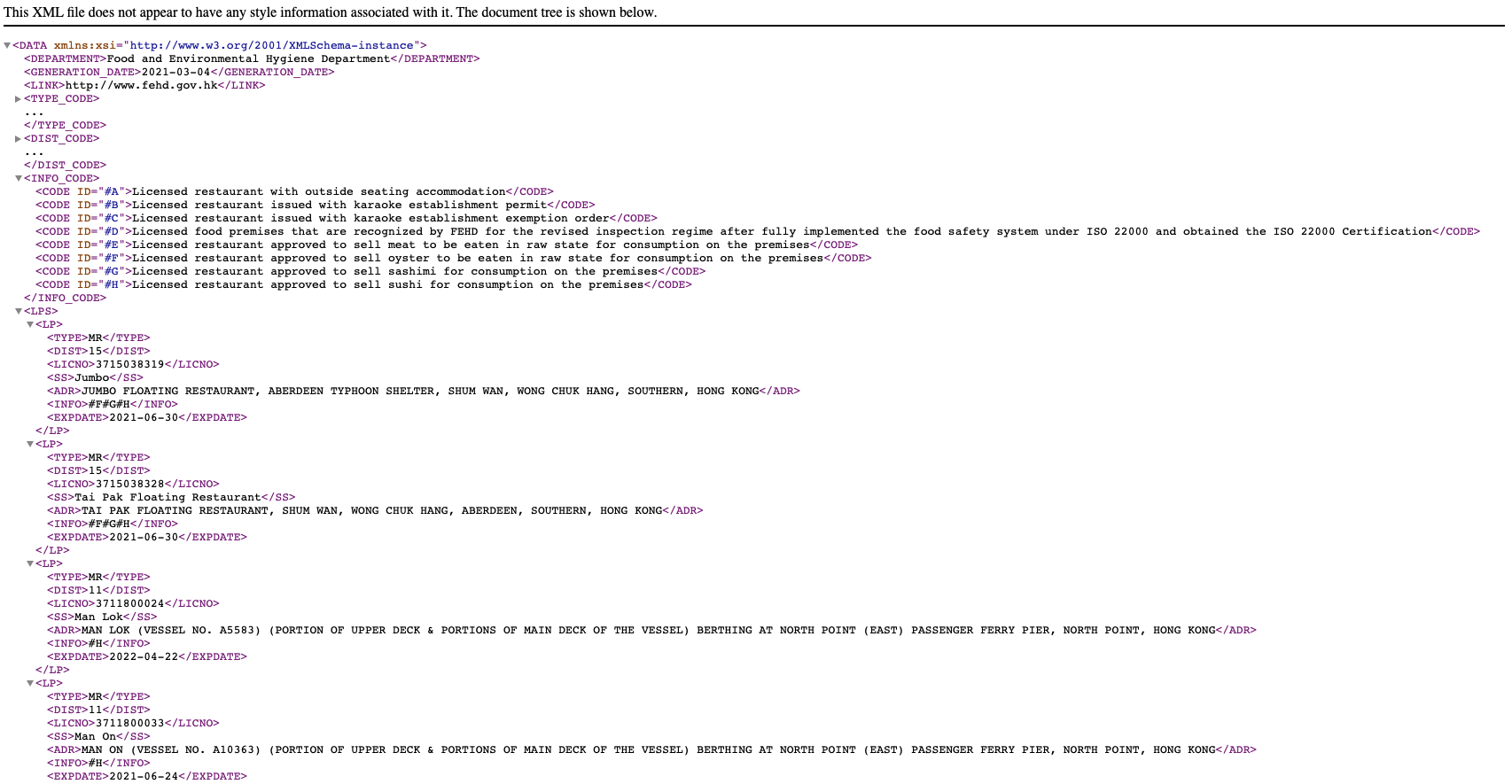
The tree structure of the XML is in the below form:
- DATA
- DEPARTMENT
- GENERATION_DATE
- LINK
- TYPE_CODE
- DIST_CODE
- INFO_CODE
- LPS
- LP
- TYPE
- DIST
- LICNO
- SS
- ADR
- INFO
- EXPDATE
- LP
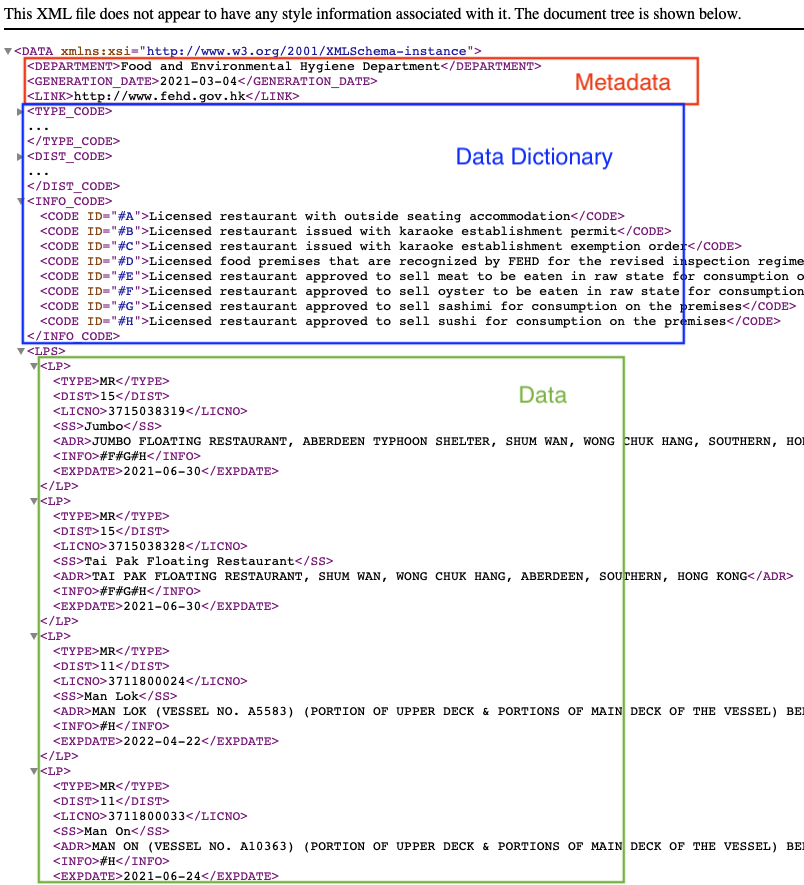
DEPARTMENT, GENERATION_DATE and LINK are metadata. TYPE_CODE, DIST_CODE and INFO_CODE are data dictionary to store the ID used to encode data. Therefore, only the parts wrapped inside the LPS is the “actual” data I need.
Transform xml to dataframe with R
library(xml2)
library(tidyverse)
First thing first, load the packages.
Transform xml to list
xml_address = "http://www.fehd.gov.hk/english/licensing/license/text/LP_Restaurants_EN.XML"
restaurant_license_xml = as_list(read_xml(xml_address))
Here read_xml from xml2 is used to read the XML file. The as_list function then turn it into an equivalent R list.
Expand the data to multiple rows by tags
xml_df = tibble::as_tibble(restaurant_license_xml) %>%
unnest_longer(DATA)
The XML becomes an extremely long list. As the XML is nested into multiple layers, the thing to do is to unnest the first layer. unnest_longer is a function in tidyr which unnest a list the split the values of the list to multiple rows (thus longer).
DATA in the parameter of unnest_longer refers to the DATA tag on the top layer of the xml. When it is unnested, the structure of the dataframe will return the data inside each tag. In addition, a new column named DATA_id is added to indicate which xml tag the data belongs to. Since we are unnesting the first layer, the DATA_id column includes these 7 values:
- DEPARTMENT
- GENERATION_DATE
- LINK
- TYPE_CODE
- DIST_CODE
- INFO_CODE
- LP
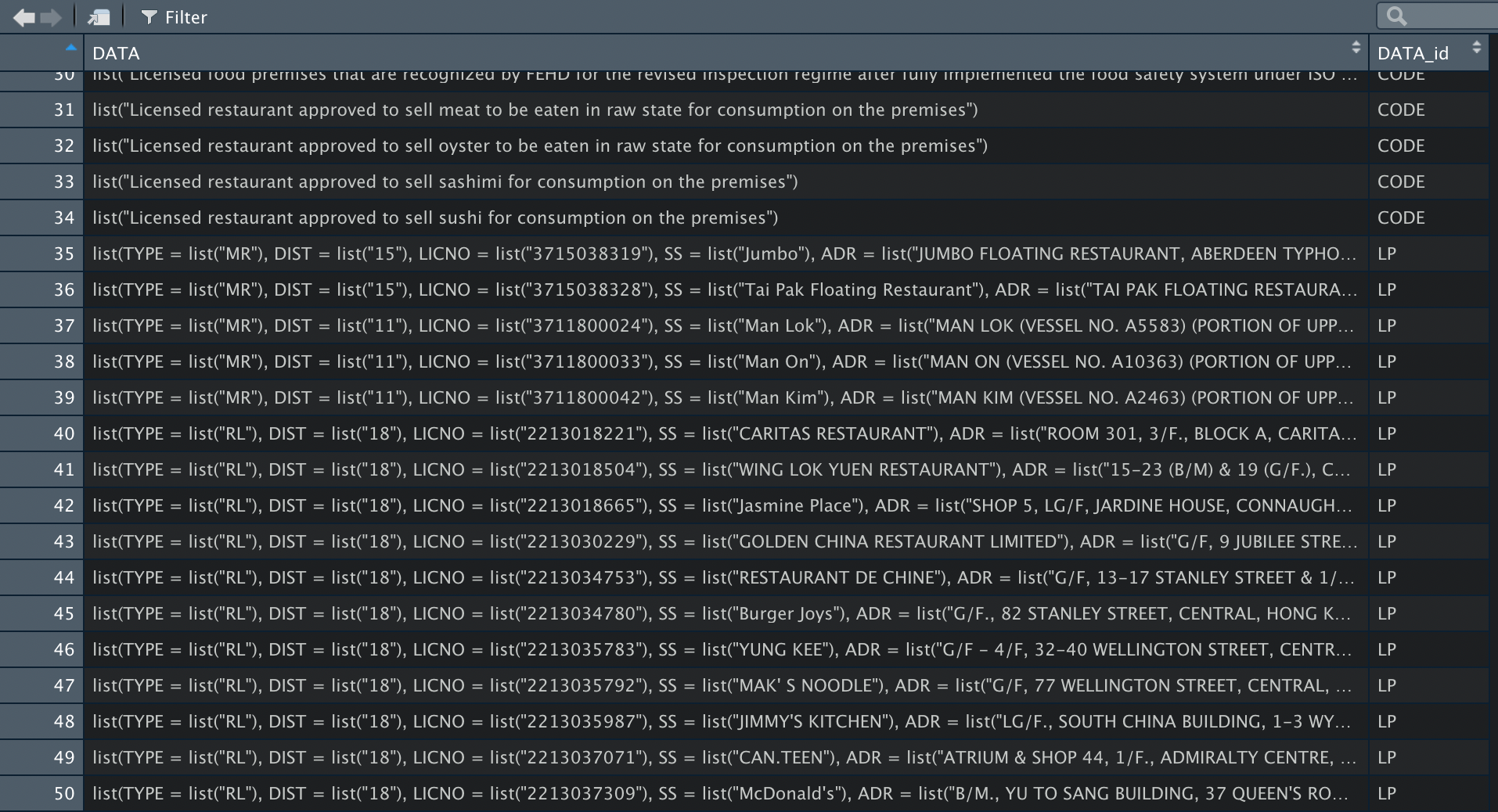
The data of each LP tag is in a list form like this:
list(
TYPE = list("MR"),
DIST = list("15"),
LICNO = list("3715038319"),
SS = list("Jumbo"),
ADR = list("JUMBO FLOATING RESTAURANT, ABERDEEN TYPHOON SHELTER, SHUM WAN, WONG CHUK HANG, SOUTHERN, HONG KONG"),
INFO = list("#F#G#H"),
EXPDATE = list("2021-06-30")
)
Compare to the XML version of the data:
<LP>
<TYPE>MR</TYPE>
<DIST>15</DIST>
<LICNO>3715038319</LICNO>
<SS>Jumbo</SS>
<ADR>JUMBO FLOATING RESTAURANT, ABERDEEN TYPHOON SHELTER, SHUM WAN, WONG CHUK HANG, SOUTHERN, HONG KONG</ADR>
<INFO>#F#G#H</INFO>
<EXPDATE>2021-06-30</EXPDATE>
</LP>
The structure of the data is thus the same. Still, it is easier to work with list in R.
Expand the data to multiple columns by tags
lp_wider = xml_df %>%
dplyr::filter(DATA_id == "LP") %>%
unnest_wider(DATA)
First, data not in the LP tags are discarded using dplyr::filter. Then, I need to expand the attributes of each license into each column (i.e. a column for TYPE, another column for DIST, etc.). Unlike the step above, this time unnest_wider is used to “expand” the data in a single column to multiple columns (thus wider).
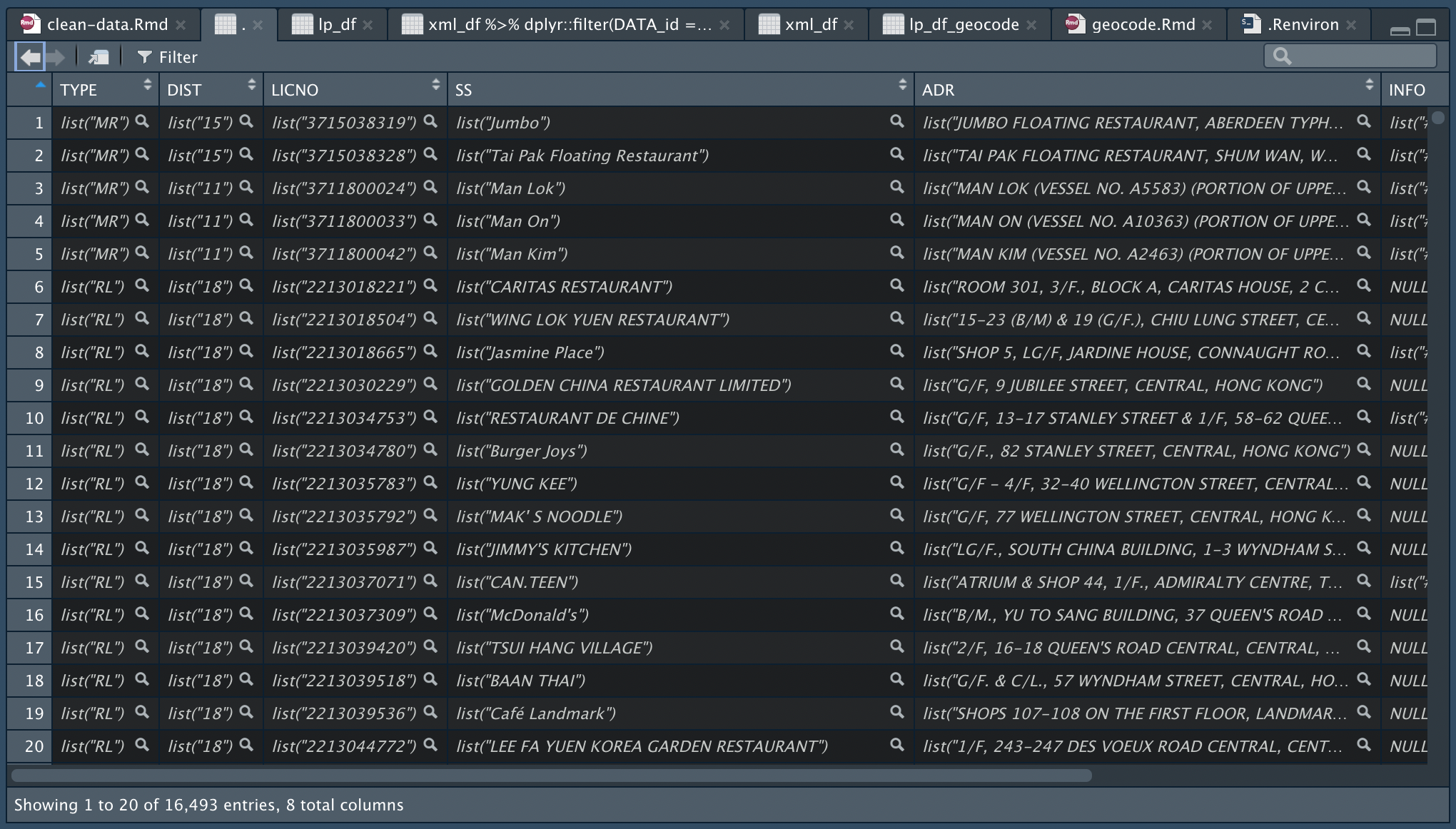
Unnest the list in each cell
lp_df = lp_wider %>%
# 1st time unnest to release the 2-dimension list?
unnest(cols = names(.)) %>%
# 2nd time to nest the single list in each cell?
unnest(cols = names(.)) %>%
# convert data type
readr::type_convert()
The data then has to be unnested two times. I do not quite understand why this is required though. Maybe because the values in the cells a in the form of “list in list”/“nested list”? Following are the results after lp_wider is passed through 1. the first unnest and 2. the second unnest. After the first unnest, each column is still a type of list with length of 1. The data are exposed only after the second unnest.
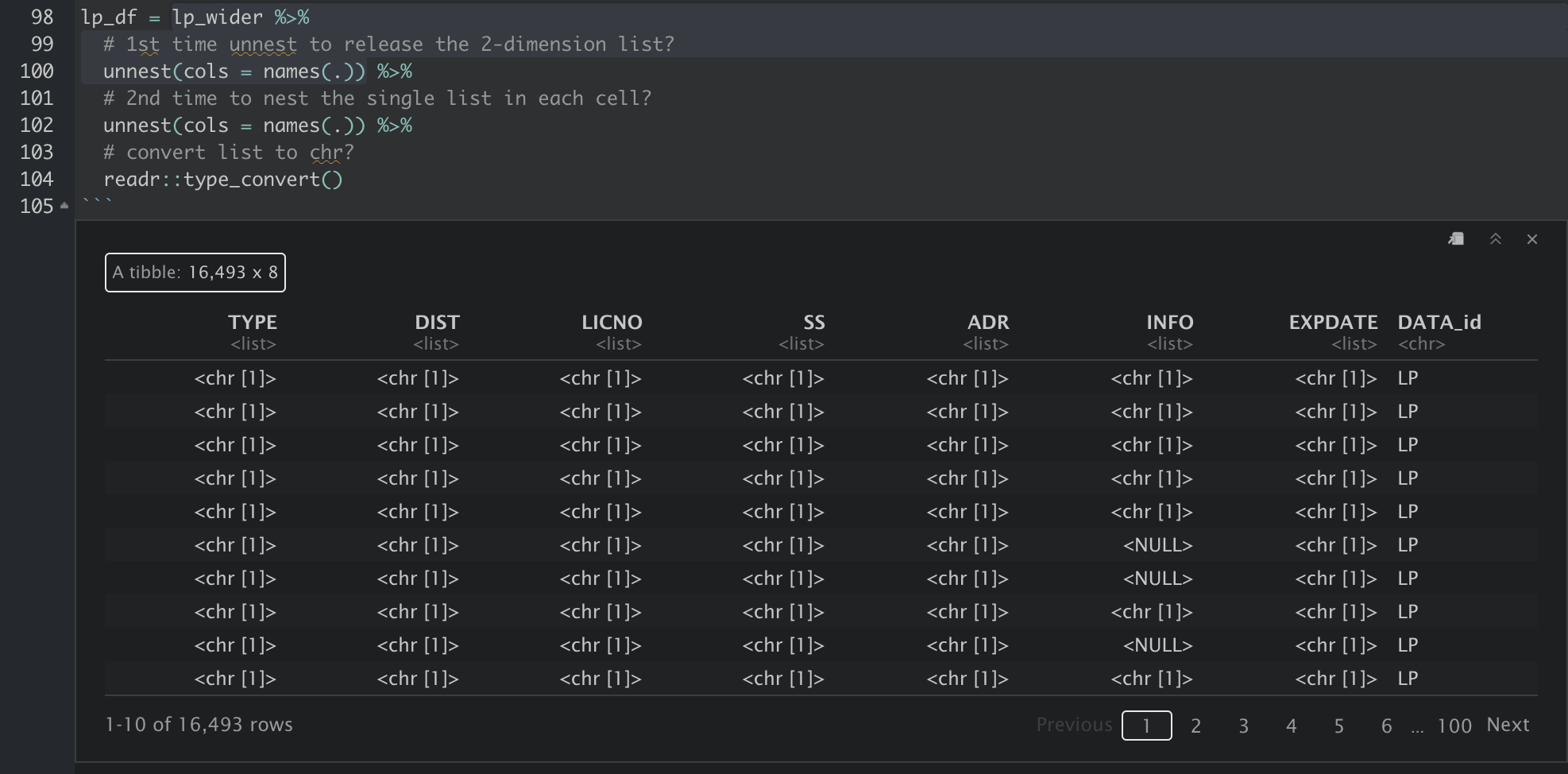
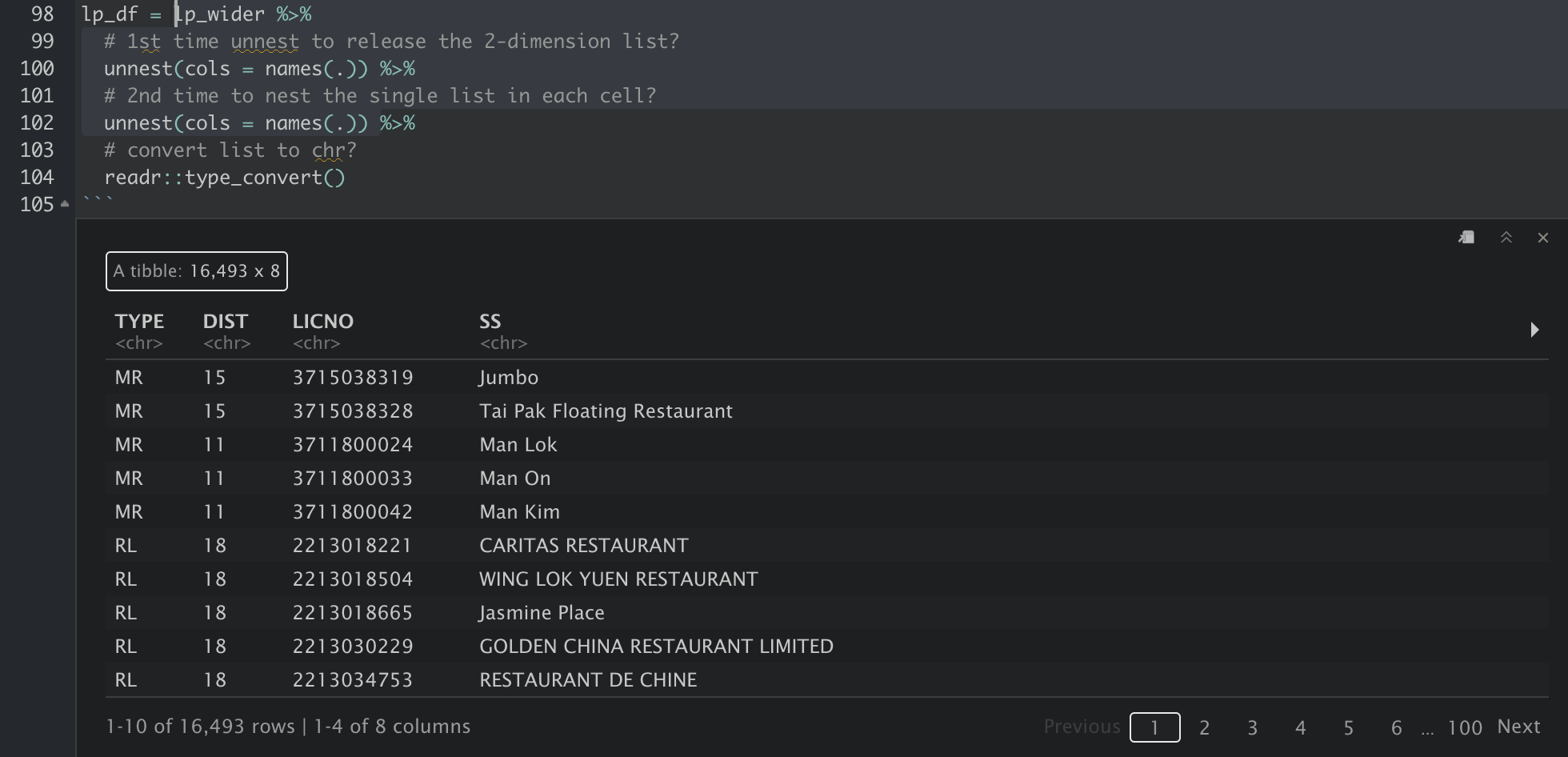
And finally, the xml is converted to a tidy tabular format for further analysis, and we can use write_csv to export the tibble into csv.
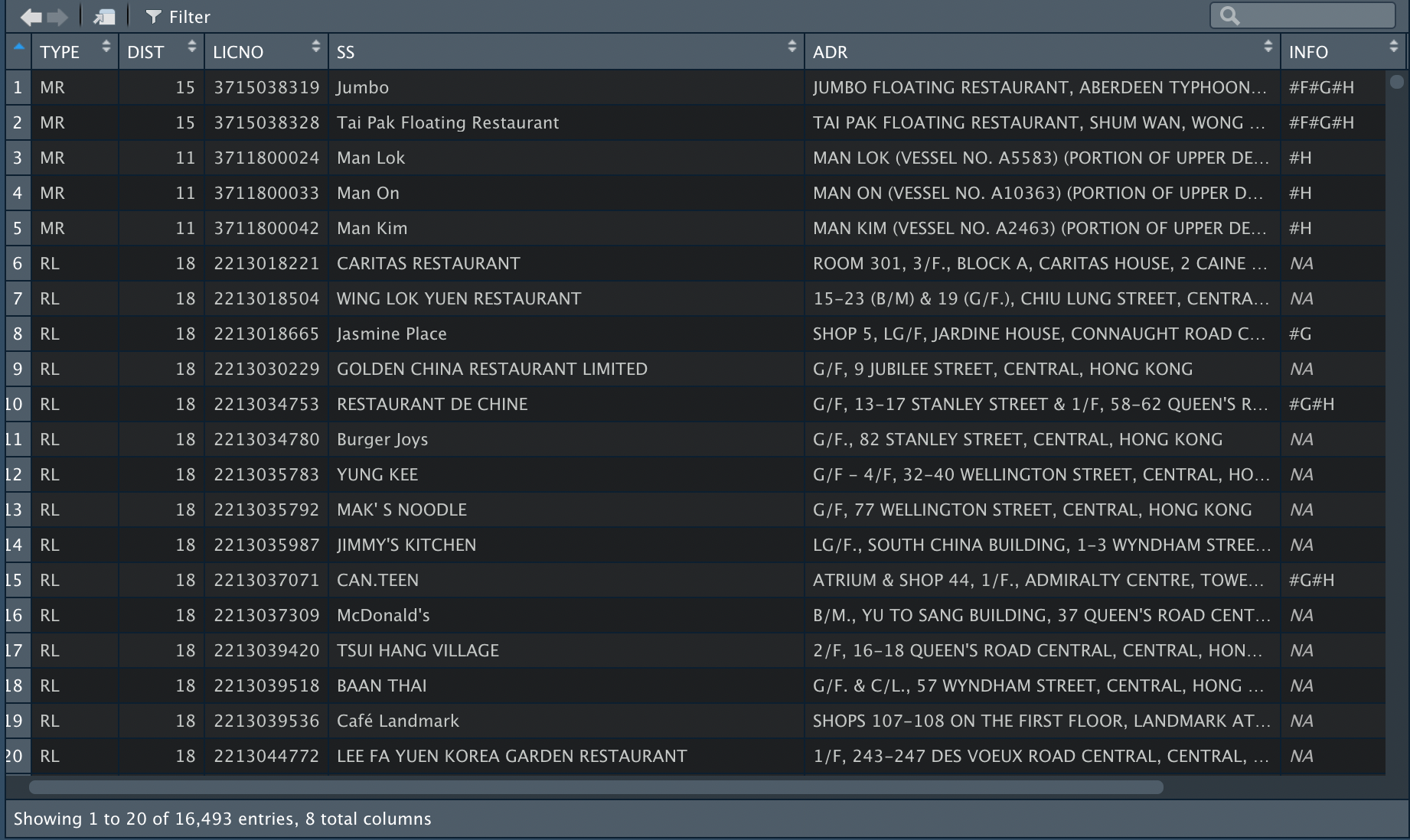
Other approaches
Manual editing and conversion
The quick (though dumb) way to get the nested part of a XML is - manually delete the “outer” part! In the case of this restaurant license XML, I only need the <LPS> part, and all other nodes (I am not sure about this is the correct terminology for XML) could be deleted. After that, I just need to use an online XML to CSV converter for the data format conversion.
Using XML package
The XML package is an older package to transform XML to dataframes.
https://blog.gtwang.org/r/r-xml-package-parsing-and-generating-xml-tutorial/
References
How to convert a partly nested XML to data frame using xml2
https://community.rstudio.com/t/how-to-convert-a-partly-nested-xml-to-data-frame-using-xml2/36705/2
nested XML to data frame in R
https://stackoverflow.com/questions/47254923/nested-xml-to-data-frame-in-r
Converting xml to tibble in R
https://megapteraphile.wordpress.com/2020/03/29/converting-xml-to-tibble-in-r/